|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Objectives of the project
One of the main goals of mapping projects is the identification and isolation of genes determining phenotypes of interest. This can be done in different ways, among which comparative genome mapping is unanimously considered the most cost-effective approach from the point of view of time and resources. This becomes especially true, as the input from genome mapping projects is progressing at an exponential rate. Comparative mapping permits transfer of information from highly studied species to species less studied with the consequent saving in time.
The objectives of the project Bridgemap are:
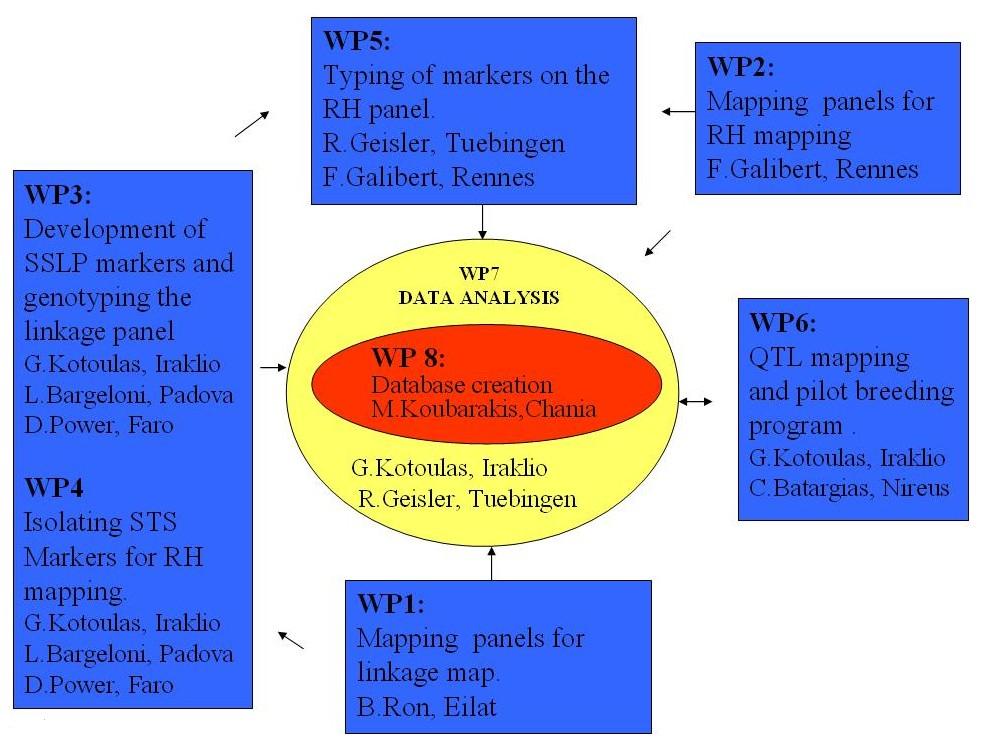
1. A relatively classical mapping approach which consists of the creation of a linkage map, towards a basic map framework that will have many applications in aquaculture as well as in population monitoring of seabream for fisheries.
2. A comparative mapping targeted approach: The project also aims to open the way for transforming gilthead seabream into a "gene rich" commercial fish species, which will serve as a model species for a rapid transfer of genetic information and tools to all marine fish species by way of comparative genomics.
3. Contribution of seabream to vertebrate genomics: a) by the production of a Radiation Hybrid map, with a linkage map with anchored genes and BAC clones for a further vertebrate species, b) by extending the genomic information available for an additional teleost species permitting a fine scale comparative analysis within fish (e.g. zebrafish versus sea bream).
The Bridgemap project started at November 2001 and its duration is 4 years.
[ Objectives | Participants | Achievements | Related Links ]
Hosted at: Technical University of Crete
Send any questions or comments regarding
this service to nfritz intelligence.tuc.gr.
intelligence.tuc.gr.